Issue 6 — Cadherins Pt. 1
Dysfunction of classical cadherins is a hallmark of cancer development and progression. They are also linked to sensorial disorders such as hereditary deafness. This piece begins the series on cadherins, allowing us to in the future assess their roles in cancer and neurobiology.
Calcium dependent mechanics of cadherin adhesion
Historical Background
The critical role of calcium ions in maintaining cell-cell adhesion strength was first established in the 1970s through the work of Masatoshi Takeichi. Using Chinese hamster (V79) cells, he demonstrated that treatment of these cells with the calcium chelator EDTA caused cell aggregates to dissociate, and that this was reversed upon the reintroduction of calcium. Importantly, dissociation was incomplete, leading Takeichi to propose that his cells must have possessed at least two adhesion mechanisms, one calcium-dependent and another calcium-independent. His subsequent embryonic tissue studies confirmed that calcium-dependent adhesion governed selective cell sorting during morphogenesis (see Takeichi, 1977, 1988). Later, his group identified and named the molecules responsible for this phenomenon, cadherins (literally Ca2+ + adherin), establishing the biochemical foundation of cell–cell adhesion. Takeichi’s work was pivotal because it extended the purely morphological observations of Townes and Holtfreter (1955) into a molecular framework and thus transformed the study of intercellular adhesion from descriptive microscopy into biochemical cell biology
Introduction
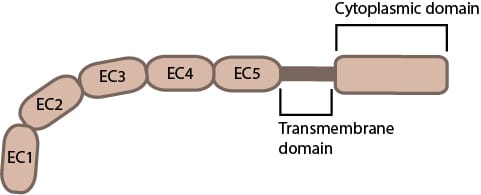
The term cadherin is today used to refer to proteins which are a part of a very broad family of receptor proteins, united by the presence of extracellular cadherin (EC) domains that mediate adhesion. There are more than 100 cadherin genes in the human genome, classified into over ten subfamilies. Broadly, cadherins are grouped as classical, desmosomal, or atypical cadherins (protocadherins, fat cadherins, etc). The distinction is based primarily on their cytoplasmic domain organisation and binding partners. Here I will briefly discuss, as an introduction to this type of proteins, the biophysics of cadherin-cadherin binding and EC domain strength of the first types of cadherins to be discovered, those which Takeichi discovered, the classical cadherins.
Binding Mechanisms of Classical Cadherins: Calcium-Dependent Rigidity and Adhesive Specificity
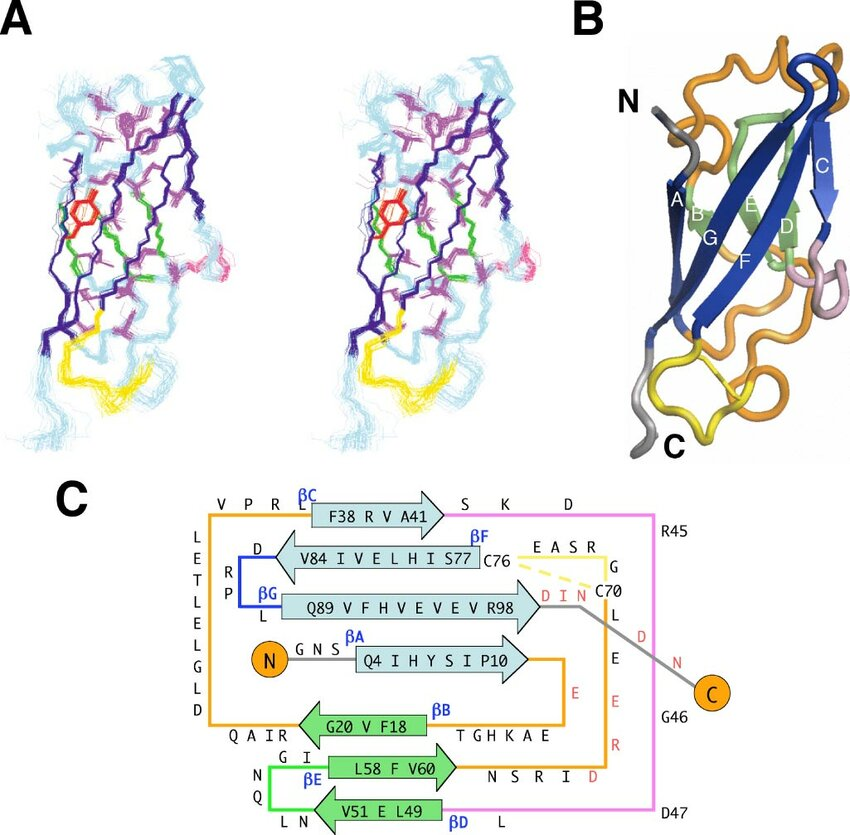
Classical type I (E-, N-, P-) and type II cadherins share a canonical ectodomain built from five tandem extracellular cadherin (EC) modules (EC1 to 5), each ~110 amino acids, organised as seven-strand β-sandwiches with Greek-key topology (see Figure 1C). The structural homology between the cadherin EC domain’s β-sandwich and the immunoglobulin (Ig) fold is of interest, to the extent that some have suggested a potential evolutionary link [1]. Be that as it may, a crucial distinction, between the two is the display and utilization by cadherins, of conserved sequence motifs that coordinate Ca²⁺ binding across inter-module linkers.
The cadherin EC module is characterized by a tightly packed hydrophobic core and an extended interstrand hydrogen-bonding network, both of which contribute to its remarkable thermodynamic stability (see figure 1A and 1B). In single-molecule force spectroscopy, unfolding forces exceeding 100 pN per cadherin domain have been recorded [2]. This value is far greater (with reason) than physiological shear stress, which for vascular endothelial cells for example ranges from roughly 1–10 pN.
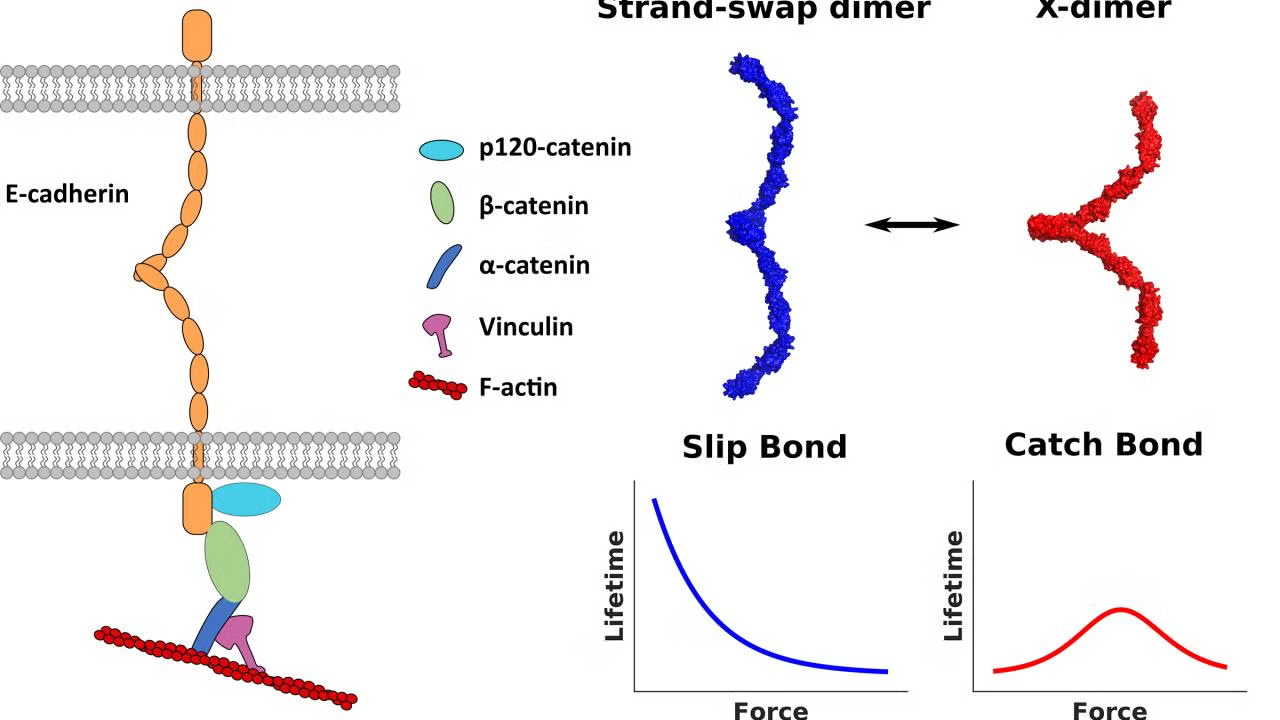
It has now been known for over 20 years that cadherin adhesive specificity is mediated by the terminal EC1 module [3] [4]. Two principal interaction geometries have been described: the strand-swapped dimer and the X-dimer (Figure 2). In the “strand-swap” dimer, the A* β-strand of one EC1 inserts its conserved Trp² into a hydrophobic pocket of its partner, forming a reciprocal domain-swapped β-sheet [5]. In the “X-dimer,” the EC1–EC2 Ca²⁺ linker participates directly in a cross-contact that pre-aligns partners for strand exchange [6]. AFM and steered molecular-dynamics (MD) experiments show that X-dimers form within milliseconds and convert to strand-swaps on the ~1-s timescale; the former behave as catch bonds (lifetime increases with modest tension, ≤ 30 pN), whereas the latter are slip bonds that dissociate under load [2]. Thus, force biases the energy landscape toward adhesive maturation.
The dependence of cadherin mediated adhesion on calcium is both chemical as calcium binds negatively charged amino acids (Asp and Glu) and structural - calcium maintains the rigidity of cadherin EC modules. The structural dependence is confirmed by high resolution crystallography (e.g., C-cadherin 1L3W; E-cadherin 3Q2V; N-cadherin 7DTY) which shows that Ca²⁺ ions bridge the C-terminal loops of one EC domain and the N-terminal loops of the next via conserved Asp/Glu residues, converting what would otherwise be a hinge into a rigid rod. Small-angle scattering and AFM confirm that the Ca²⁺-loaded ectodomain has a persistence length of ≈ 40 nm, consistent with its function as a tension-bearing lever between opposing membranes [7] [8].
Additionally, at the nanoscale, Ca²⁺ binding and inter-domain rigidity act as mechanical filters. Molecular dynamics simulations show that Ca²⁺ removal increases linker flexibility by ≈ 15°, reducing effective end-to-end stiffness and lowering unfolding forces by > 40 % [9]. Extensive hydrogen-bonding between β-strands A and G spans the hydrophobic core, generating long-range cooperativity within the domain. This cooperativity promotes sequential, domain-wise unfolding under tension, observed in AFM trajectories as characteristic 35-nm extension steps. This sequential rupture distributes load and prevents catastrophic detachment, explaining how cadherin junctions sustain chronic shear yet remodel rapidly when Ca²⁺ is depleted or proteases (e.g., HtrA from H. pylori) cleave linker loops [10].
Calcium coordination therefore dictates both adhesion strength and protease resistance. Each inter-domain Ca²⁺ triad binds with a moderate affinity (Kd ≈ 10⁻⁵ M). This Kd means that micromolar fluctuations in Ca²⁺ concentrations, e.g. near wound edges or synapses, can rapidly modulate ectodomain stiffness and clustering without fully dismantling the junction. This rigidity translates directly into cell-level mechanical strength. In epithelia, Ca²⁺ chelation by EGTA reduces overall junctional tension by ~70 %, (measured via FRET-based tension sensors) [11]. When stiffened, these EC arrays efficiently transmit mechanical load across the cell membrane chanelling ~ 2-5 pN per dimer to the cytoplasmic actin anchor a finding which matches in-cell measurements [12].
References
1. Overduin, M., et al., Solution Structure of the Epithelial Cadherin Domain Responsible for Selective Cell Adhesion. Science, 1995. 267(5196): p. 386-389.
2. Rakshit, S., et al., Ideal, catch, and slip bonds in cadherin adhesion. Proceedings of the National Academy of Sciences, 2012. 109(46): p. 18815-18820.
3. Boggon, T.J., et al., C-Cadherin Ectodomain Structure and Implications for Cell Adhesion Mechanisms. Science, 2002. 296(5571): p. 1308-1313.
4. Pertz, O., A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. The EMBO Journal, 1999. 18(7): p. 1738-1747.
5. Harrison, O.J., et al., Two-step adhesive binding by classical cadherins. Nature Structural & Molecular Biology, 2010. 17(3): p. 348-357.
6. Brasch, J., et al., Thinking outside the cell: how cadherins drive adhesion. Trends in Cell Biology, 2012. 22(6): p. 299-310.
7. Pokutta, S., et al., Conformational changes of the recombinant extracellular domain of E‐cadherin upon calcium binding. European Journal of Biochemistry, 1994. 223(3): p. 1019-1026.
8. Cailliez, F. and R. Lavery, Cadherin Mechanics and Complexation: The Importance of Calcium Binding. Biophysical Journal, 2005. 89(6): p. 3895-3903.
9. Tiwari, P., et al., Structural-Mechanical and Biochemical Functions of Classical Cadherins at Cellular Junctions: A Review and Some Hypotheses. 2018, Springer Singapore. p. 107-138.
10. Schmidt, T.P., et al., Identification of E-cadherin signature motifs functioning as cleavage sites for Helicobacter pylori HtrA. Scientific Reports, 2016. 6(1): p. 23264.
11. Borghi, N., et al., E-cadherin is under constitutive actomyosin-generated tension that is increased at cell–cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences, 2012. 109(31): p. 12568-12573.
12. Benham-Pyle, B.W., B.L. Pruitt, and W.J. Nelson, Mechanical strain induces E-cadherin–dependent Yap1 and β-catenin activation to drive cell cycle entry. Science, 2015. 348(6238): p. 1024-1027.